All molecules display alternative conformations which show up in probabilities proportional to their Boltzmann weight. So, if we know the interactions between the atoms of a molecule (and its environment if it is in a solvent), we can calculate with relative ease, roughly how many of each conformer will appear in any given environment. However, interesting properties are usually dynamical, i.e., one might be interested in which conformer is accessible from a given one within a predetermined time range. This approach requires knowledge of the kinetics of the process and is a much harder problem to solve.
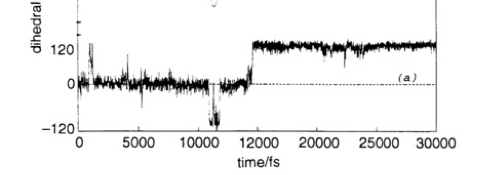
In this project, we will approach this problem with a simple system. We will use polyethylene (PE), a chain of CH2 units repeating over and over again. It is known that each of the dihedral (also called torsional) angles along the backbone is in one of three conformers, trans (planar), gauche+ or gauche- (120 degrees rotated into and out of the plane, respectively). Let’s label these t, g+ and g-. We ask the question, given a chain of conformation can we classify what the conformation of a selected position along this chain will be within a selected time frame, e.g., given a polymer is in the …ttg-tt… conformer at time 10300 fs, what will the conformation be at time 11000 fs? (see figure at the bottom for the trajectory of the central dihedral angle; from Baysal et al, J. Chem. Soc. Faraday Trans., 1995).
We will attack this problem using machine learning. We will generate a large set of plausible conformations of PE using classical molecular dynamics (MD) simulations, and we will learn from the many geometrical features of these conformers. Our first approach will be to use simple classification models. We will then increase the level of complexity of the models as we better understand the problem and its possible solutions.The findings will have direct applications in protein conformation, dynamics and function.
In this project, you will learn to run MD simulations on open-source software, and you will write your own analysis codes in Tcl and Python. The project will also require you to work with large amounts of data, consisting mainly of the coordinates of the systems.
About Project Supervisors
Canan Atilgan
canan@sabanciuniv.edu
http://people.sabanciuniv.edu/canan/
Ali Rana Atilgan
atilgan@sabanciuniv.edu
http://people.sabanciuniv.edu/atilgan/
.jpg)